Peer-reviewed research
Bold author names are FERG members. Asterisks denote equal authorship.
2025:
South, Sabini, Pattison & Cuff (2025). Aquatic biological invasions exacerbate nutritional and health inequities. Trends in Ecology and Evolution, in press.
Wilder, Herzog, Reeves, Knowles & Cuff (2025). Bridging digestive physiology and ecology for a more integrative understanding of invertebrate predators. Journal of Experimental Biology, 228 (14): jeb249697.
Tercel, Cuff, Symondson, Moorhouse-Gann, Bishop, Cole, Jolin, Govier, Chambon, Mootoocurpen, Goder & Vaughan (2025). Threatened endemic arthropods and vertebrates partition their diets with non-native ants in an isolated island ecosystem. Ecology, 106(7): e70158.
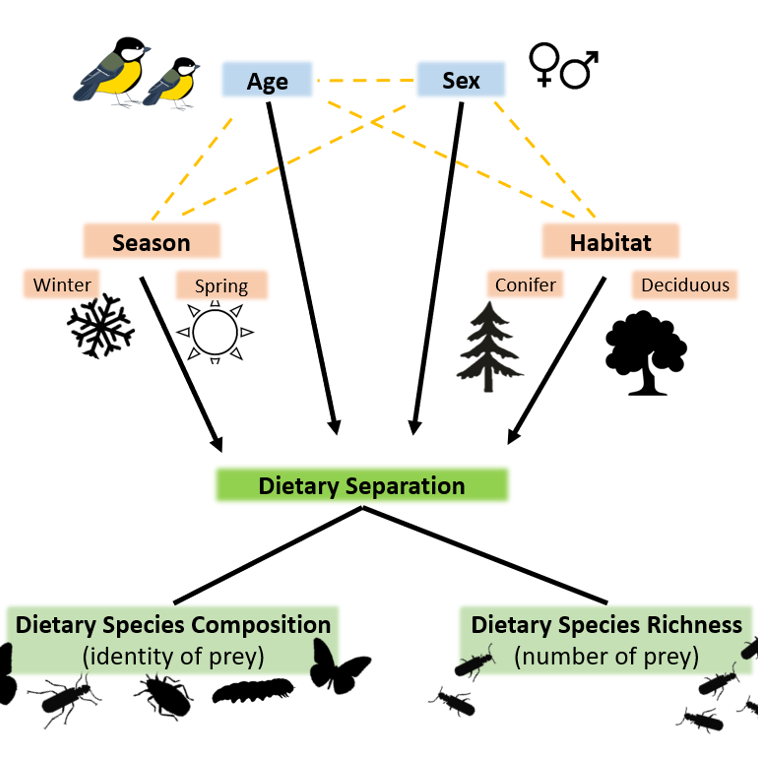
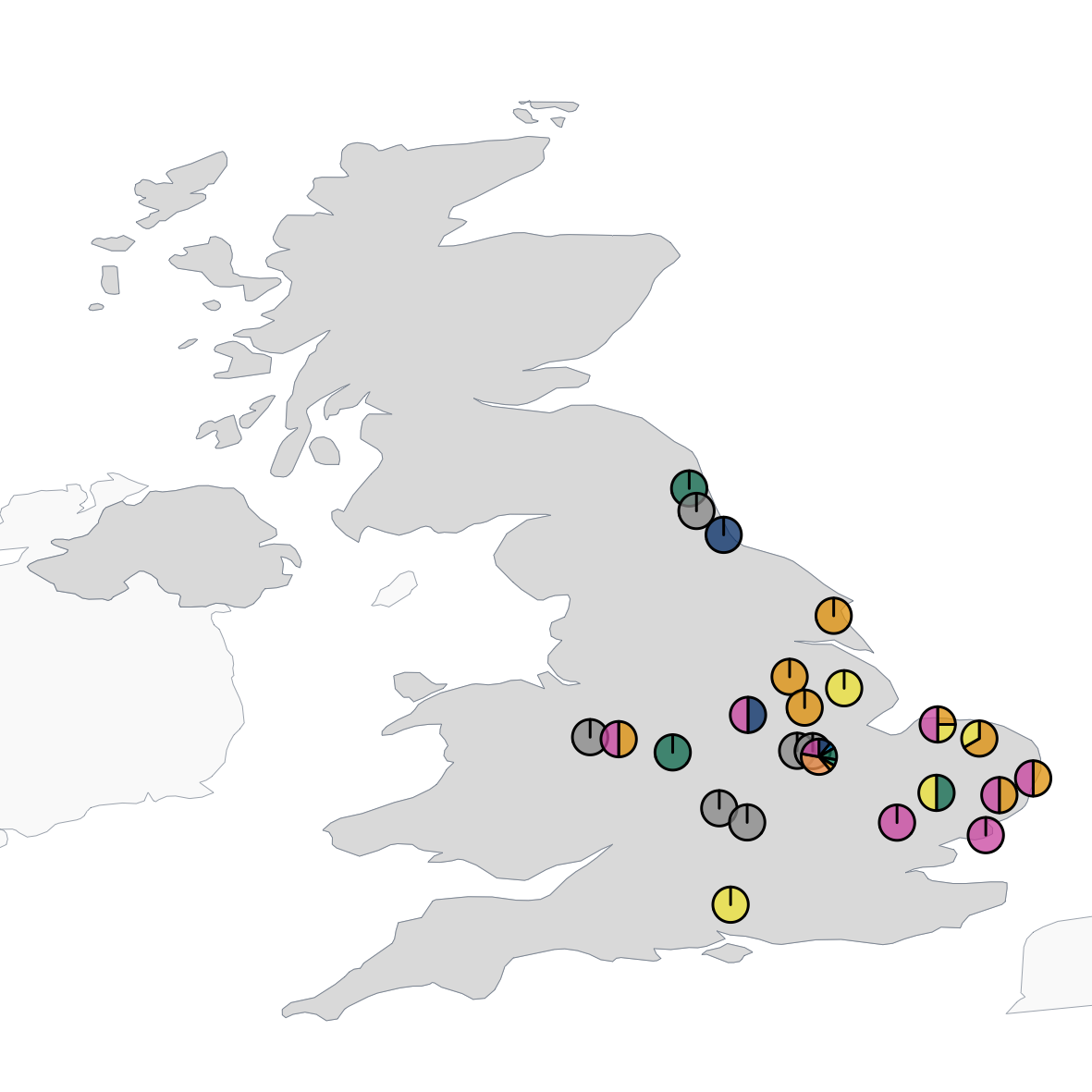
Coomes, Cuff, Reichert, Davidson, Symondson & Quinn (2025). Spatio-temporal variation in diet among age and sex cohorts of a model generalist bird species, the great tit Parus major: new insights revealed by DNA metabarcoding. Ecology and Evolution, 15 (7): e71565.
Ong, Pinoy, Lim, Bjerge, Peris-Felipo, Lind, Cuff, Cook & Høye (2025). ScannerVision: Scanner-based image acquisition of medically important arthropods for the development of computer vision and deep learning models. Current Research in Parasitology & Vector-Borne Diseases, 7: 100268.
Cuff, Tercel, Vaughan, Drake, Wilder, Bell, Müller, Orozco-terWengel & Symondson (2025). Prey nutrient content is associated with the trophic interactions of spiders and their prey selection under field conditions. Oikos, 2025 (2): e10712.
Cuff & Watt (2025). Advances in insect biomonitoring for agriculture and forestry: A synthesis on a multifaceted special issue of Agricultural and Forest Entomology. Agricultural and Forest Entomology, 27 (1): 1-7.
Cuff, Gajski, Michalko, Košulič & Pekár (2025). Biomonitoring of biocontrol across the full annual cycle in temperate climates: post-harvest, winter and early-season interaction data and methodological considerations for its collection. Agricultural and Forest Entomology, 27 (1): 18-34.
Hawthorne, Cuff, Collins & Evans (2025). Metabarcoding advances agricultural invertebrate biomonitoring by enhancing resolution, increasing throughput, and facilitating network inference. Agricultural and Forest Entomology, 27 (1): 50-66.
2024:
Cuff, Evans, Vaughan, Wilder, Tercel & Windsor (2024). Networking nutrients: how nutrition determines the structure of ecological networks. Journal of Animal Ecology, 93 (8): 974-988.
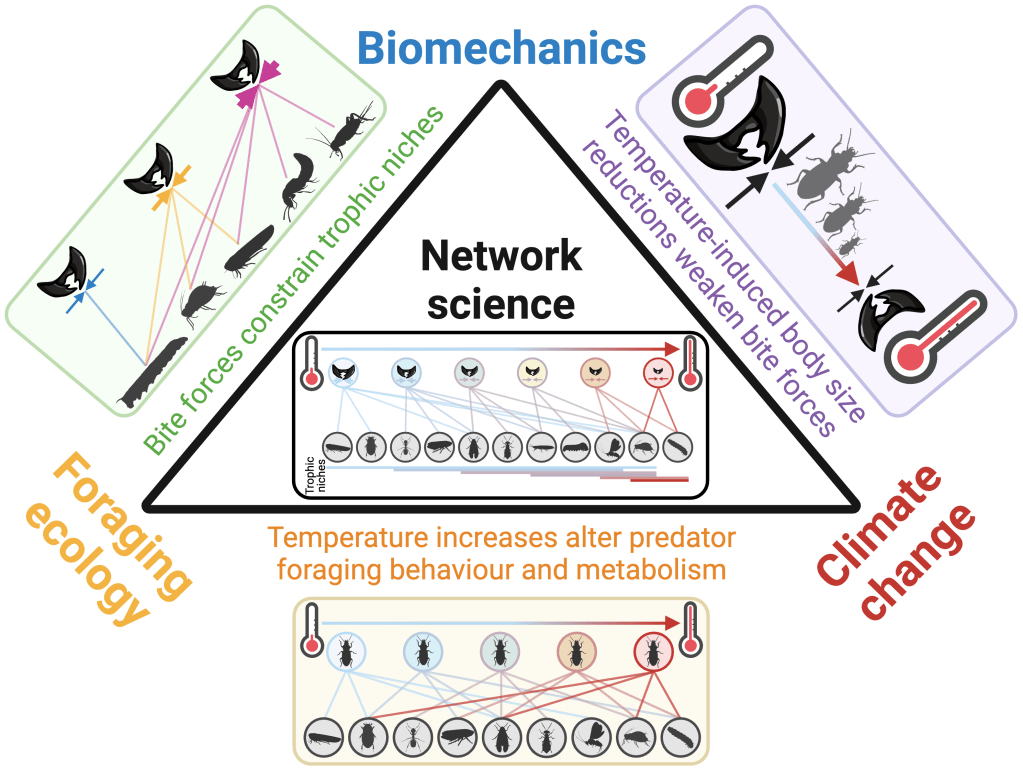
Cuff, Labonte & Windsor (2024). Understanding trophic interactions in a warming world by bridging foraging ecology and biomechanics with network science. Integrative and Comparative Biology, 64 (2): 306-321.
Tercel, Cuff, Vaughan, Symondson, Goder, Matadeen, Tatayah & Cole (2024). Ecology, natural history, and conservation status of Scolopendra abnormis, a threatened centipede endemic to Mauritius. Endangered Species Research, 54: 181-189.
Petsopoulos, Cuff, Bell, Kitson, Collins, Boonham, Morales-Hojas & Evans (2024). Identifying archived insect bulk samples using DNA metabarcoding: a case study using the long-term Rothamsted Insect Survey. Environmental DNA, 6 (3): e542.
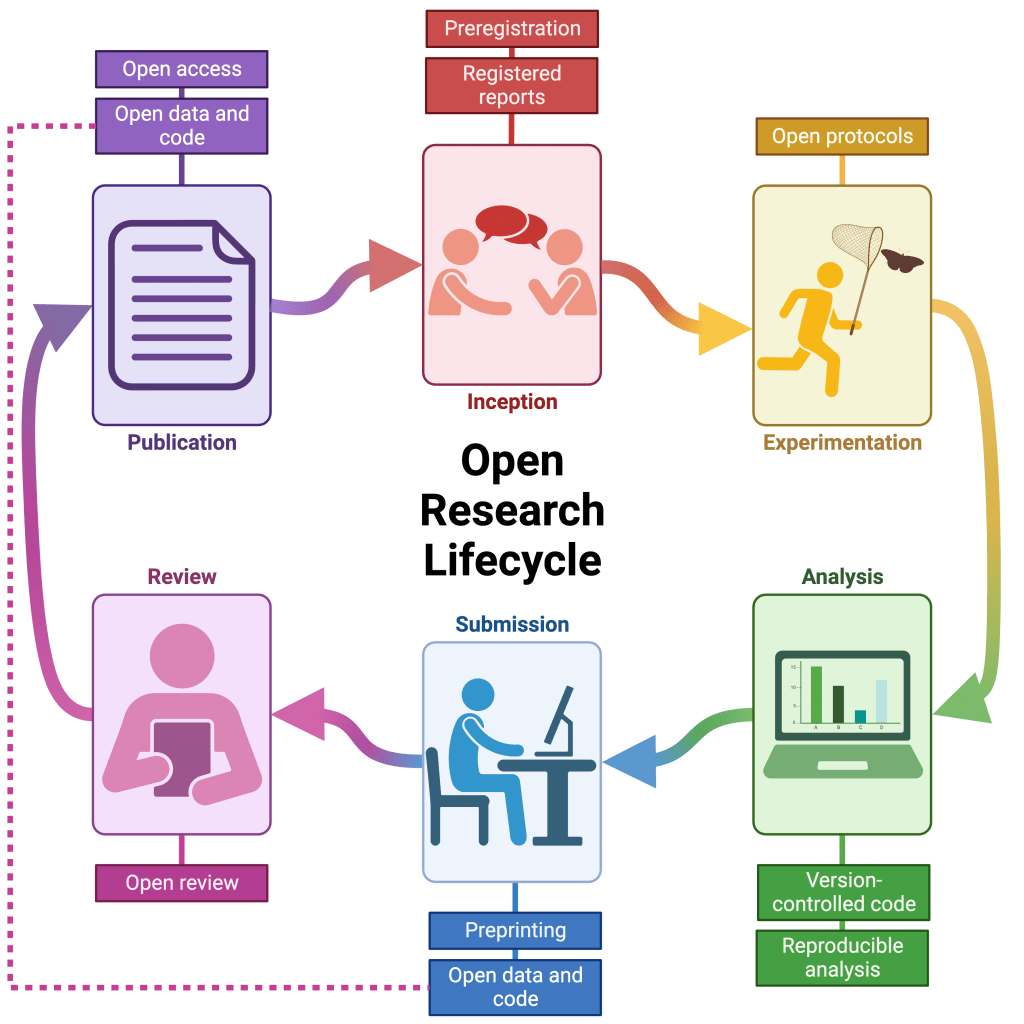
Cuff, Barrett, Gray, Fox, Watt & Aimé (2024). The case for open research in entomology: reducing harm, refining reproducibility and advancing insect science. Agricultural and Forest Entomology, 26 (3): 285-295.
Cuff, Tercel, Windsor, Hawthorne, Hambäck, Bell, Symondson & Vaughan (2024). Sources of prey availability data alter interpretation of outputs from prey choice null networks. Ecological Entomology, 49 (3): 418-432.
2023:
Cuff, Dighe, Watson, Badell-Grau, Weightman, Jones & Kille (2023). Monitoring SARS-CoV-2 using infoveillance, national reporting data and wastewater in Wales, UK. JMIR Infodemiology, 3: e43891.
Tercel, Cuff, Symondson & Vaughan (2023). Non-native ants drive dramatic declines in animal community diversity: A meta-analysis. Insect Conservation and Diversity, 16 (6): 733-744.
Cuff, Windsor, Tercel, Bell, Symondson & Vaughan (2023). Temporal variation in spider trophic interactions is explained by the influence of weather on prey communities, web building and prey choice. Ecography, 2023 (7): e06737.
Drake, Cuff, Bedmar, McDonald, Symondson & Chadwick (2023). Otterly delicious: Spatiotemporal variation in the diet of a recovering population of Eurasian otters (Lutra lutra) revealed through DNA metabarcoding and morphological analysis of prey remains. Ecology and Evolution, 13 (5): e10038.
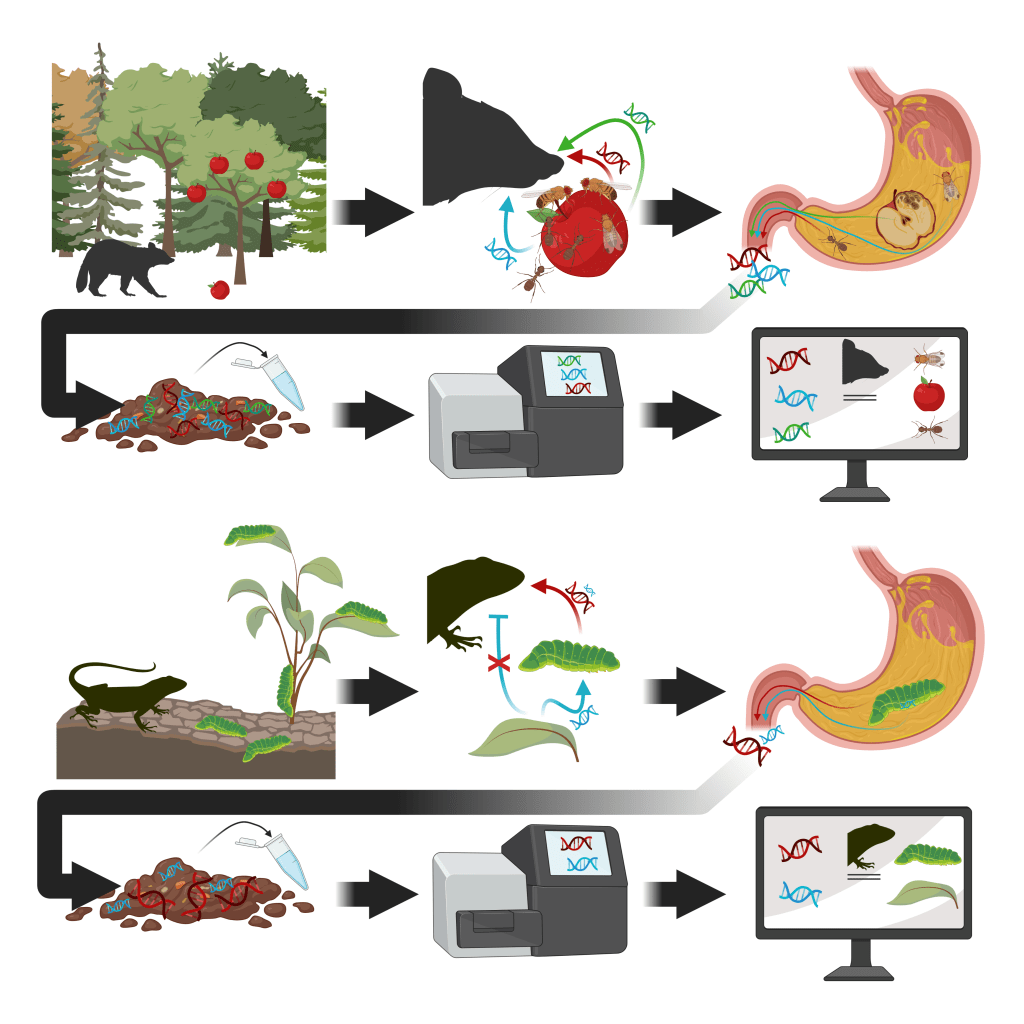
Cuff, Kitson, Hemprich-Bennett, Tercel, Browett & Evans (2023). The predator problem and PCR primers in molecular dietary analysis: Swamped or silenced; depth or breadth? Molecular Ecology Resources, 23 (1): 41-51.
2022:
Tercel* & Cuff* (2022). The complex epistemological challenge of data curation in dietary metabarcoding: Comment on Littleford-Colquhoun et al. (2022). Molecular Ecology, 31 (22): 5653-5659.
Cuff, Evans, Porteous, Quiñonez & Evans (2022). Candy-striped spider leaf and habitat preferences for egg deposition. Agricultural and Forest Entomology, 24 (3): 422-431.
Cuff, Windsor, Tercel, Kitson & Evans (2022). Overcoming the pitfalls of merging dietary metabarcoding into ecological networks. Methods in Ecology and Evolution, 13 (3): 545-559.
Tercel, Moorhouse-Gann, Cuff, Drake, Cole, Goder, Mootoocurpen & Symondson (2022). DNA metabarcoding reveals introduced species predominate in the diet of a threatened endemic omnivore, Telfair’s skink (Leiolopisma telfairii). Ecology and Evolution, 12 (1): e8484.
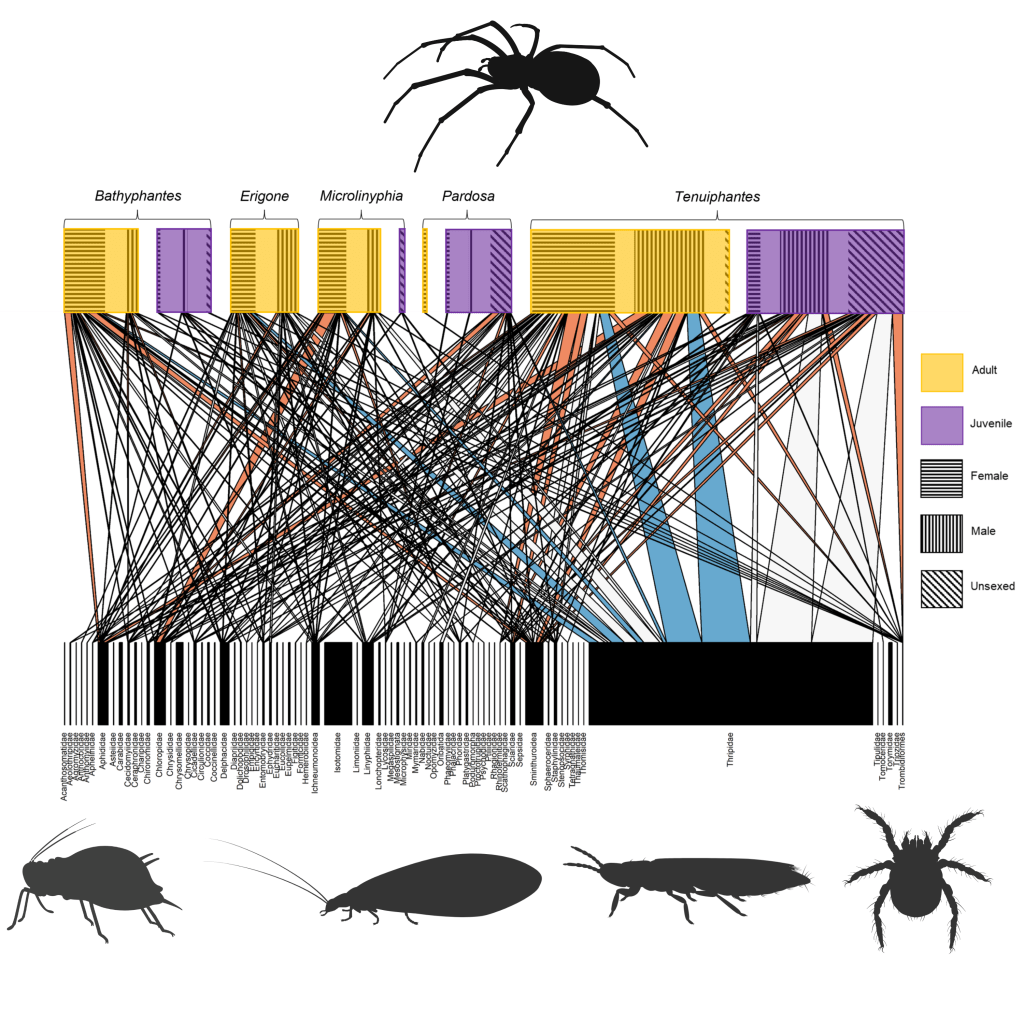
Cuff, Tercel, Drake, Vaughan, Bell, Orozco-terWengel, Müller & Symondson (2022). Density-independent prey choice, taxonomy, life history, and web characteristics determine the diet and biocontrol potential of spiders (Linyphiidae and Lycosidae) in cereal crops. Environmental DNA, 4 (3): 549-564.
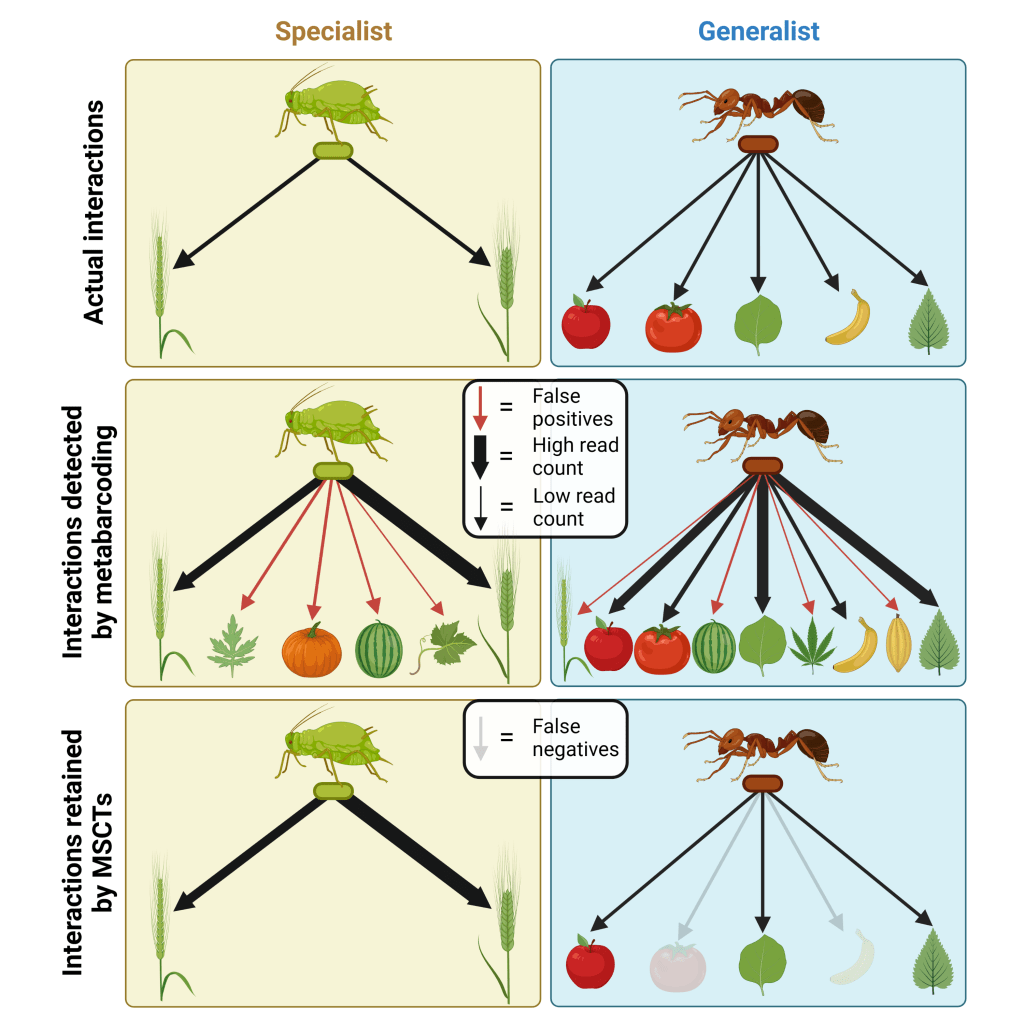
Drake, Cuff, Young, Marchbank, Chadwick & Symondson (2022). An assessment of minimum sequence copy thresholds for identifying and reducing the prevalence of artefacts in dietary metabarcoding data. Methods in Ecology and Evolution, 13 (3): 694-710.
2021:
Cuff, Aharon, Steinpress, Seifan, Lubin & Gavish-Regev (2021). It’s all about the zone: spider assemblages in different ecological zones of Levantine caves. Diversity, 13 (11): 576.
Cuff*, Windsor*, Gilmartin, Boddy & Jones (2021). Influence of European beech (Fagales: Fagaceae) rot hole habitat characteristics on invertebrate community structure and diversity. Journal of Insect Science, 21 (5): 7.
Tercel, Symondson & Cuff (2021). The problem of omnivory: A synthesis on omnivory and DNA metabarcoding. Molecular Ecology, 30 (10): 2199-2206.
Cuff, Wilder, Tercel, Hunt, Oluwaseun, Morley, Badell-Grau, Vaughan, Bell, Orozco-terWengel, Symondson & Müller (2021). MEDI: Macronutrient Extraction and Determination from invertebrates, a rapid, cheap and streamlined protocol. Methods in Ecology and Evolution, 12 (4): 593-601.
Cuff, Drake, Tercel, Stockdale, Orozco-terWengel, Bell, Vaughan, Müller & Symondson (2021). Money spider dietary choice in pre- and post-harvest cereal crops using metabarcoding. Ecological Entomology, 46 (2): 249-261.
2020:
Badell-Grau*, Cuff*, Kelly, Waller-Evans, Lloyd-Evans (2020). Investigating the prevalence of reactive online searching in the COVID-19 pandemic: infoveillance study. Journal of Medical Internet Research, 22 (10): e19791.
Cuff, Müller, Gilmartin, Boddy & Jones (2020). Home is where the heart rot is: violet click beetle, Limoniscus violaceus (Müller, 1821), habitat attributes and volatiles. Insect Conservation and Diversity, 14 (1): 155-162.
Ammann, Moorhouse-Gann, Cuff, Bertrand, Mestre, Hidalgo, Ellison, Herzog, Entling, Albrecht & Symondson (2020). Insights into aphid prey consumption by ladybirds: optimising field sampling methods and primer design for high throughput sequencing. Plos One, 15 (7): e0235054.
Telfer, Cuff, Spelda & Owen (2020). Neobisium simile (L. Koch, 1873) (Pseudoscorpiones: Neobisiidae): a species new to Britain. Arachnology, 18 (5): 490-496.
Lafage, Elbrecht, Cuff, Steinke, Hambäck, Erlandsson (2020). A new primer for metabarcoding of spider gut contents. Environmental DNA, 2 (2): 234-243.
2019:
Bajwa, Cuff, Imran, Islam, Mansha, Ashraf, Khan, Rashid, Zahoor, Khan, Rehman, Nadeem, Orozco-terWengel & Shehzad (2019). Assessment of nematodes in Punjab Urial (Ovis vignei punjabiensis) population in Kalabagh Game Reserve: development of a DNA barcode approach. European Journal of Wildlife Research, 65: 63.
Books and book sections

Cuff (2024). Ecology. Insects. Edited by: Roy, Hill, Watt & Aimé.
Cuff, Dopson*, Hawthorne*, Howells*, Kitson*, Miller*, Suresh*, Xin*, Evans (2023). A roadmap for biomonitoring in the 21st century: merging methods into metrics via ecological networks. Advances in Ecological Research, 68: 1-34.
Cuff (2020). ‘Using DNA metabarcoding to analyse the gut contents of spiders’ in Wheater et al. Practical Field Ecology. 2nd edition, Wiley-Blackwell, pp 121-124.
Preprints
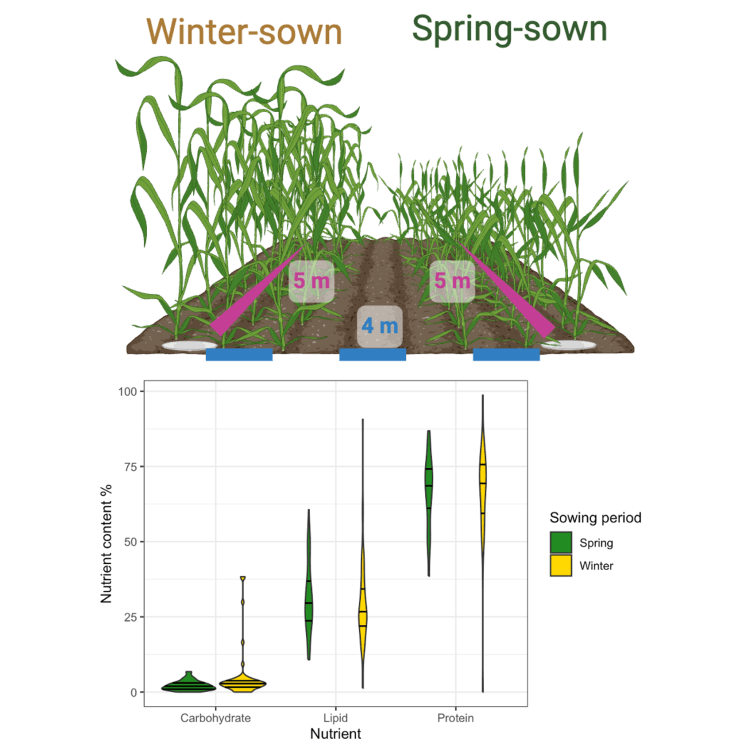
Christopher*, Wright*, Windsor & Cuff (2025). Arthropod predator nutrient content changes with crop sowing period with implications for biocontrol. bioRxiv.
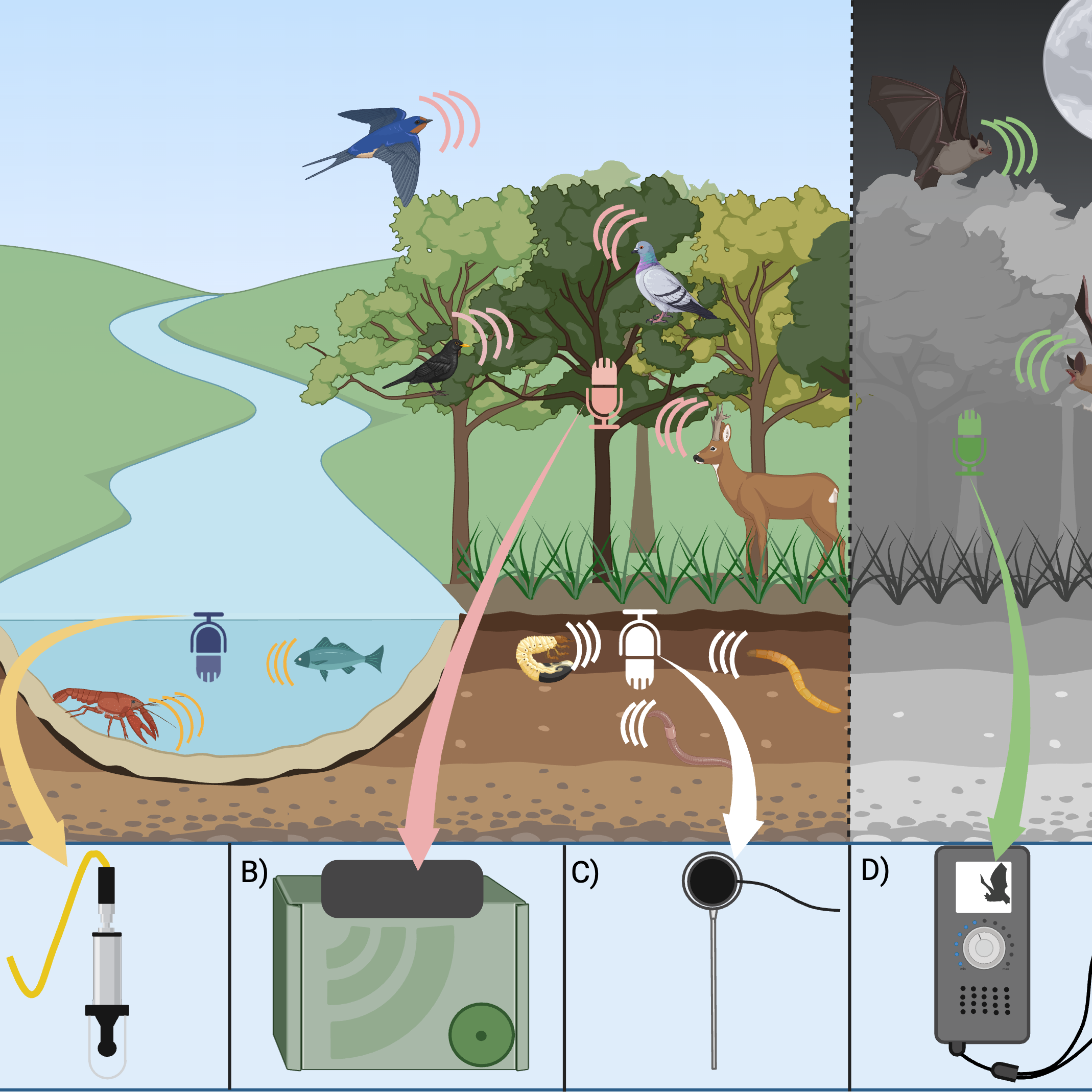
Dawson, Evans, Abrahams, Kitson, Collins & Cuff (2025). Ecoacoustics for context-rich direct and indirect trophic interaction data and ecological network construction. OSF Preprints.
Cuff, Kitson, Windsor, Sint, Hawthorne, Traugott, Anderson, Neidel, Gatehouse, Thwaites, Edwards & Evans (2025). Dietary RNA: diagnostic and functional RNA offer a potential paradigm shift for molecular dietary analyses. OSF Preprints.
Coomes, Cuff, Reichert, Davidson, Symondson & Quinn (2024). Spatio-temporal variation in diet among age and sex cohorts of a model generalist bird species, the great tit Parus major, revealed by DNA metabarcoding. Authorea.
Attar, Kitson, Cuff, Howard, Lages, Gomez & Boonham (2024). Identifying the Fusarium species involved in foot rot disease of faba beans in the UK using a combined molecular and microbiological approach. biorxiv.
Cuff, Gajski, Michalko, Košulič & Pekár (2024). Biomonitoring of biocontrol across the full annual cycle in temperate climates: post-harvest, winter and early-season interaction data and methodological considerations for its collection. OSF Preprints.
Cuff, Labonte & Windsor (2023). Understanding trophic interactions in a warming world by bridging foraging ecology and biomechanics with network science. OSF Preprints.
Hawthorne, Cuff, Collins & Evans (2023). Metabarcoding advances agricultural invertebrate biomonitoring by enhancing resolution, increasing throughput, and facilitating network inference. OSF Preprints.
Cuff, Barrett, Gray, Fox, Watt & Aimé (2023). The case for open research in entomology: reducing harm, refining reproducibility and advancing insect science. OSF Preprints.
Cuff, Tercel, Windsor, Hawthorne, Hambäck, Bell, Symondson & Vaughan (2023). Sources of prey availability data alter interpretation of outputs from prey choice null networks. bioRxiv.
Drake, Cuff, Bedmar, McDonald, Symondson & Chadwick (2023). Otterly delicious: Spatiotemporal variation in the diet of a recovering population of Eurasian otters (Lutra lutra) revealed through DNA metabarcoding and morphological analysis of prey remains. Authorea.
Cuff, Windsor, Tercel, Bell, Symondson & Vaughan (2022). Weather modulates spider trophic interactions: the interactive effects of prey community structure, adaptive web building and prey choice. Authorea.
Cuff et al. (2022). An infoveillance analysis of public interest, national data and wastewater monitoring in Wales, UK. JMIR Preprints.
Cuff et al. (2022). Networking nutrients: how nutrition determines the structure of ecological networks. Authorea.
Cuff et al. (2022). The predator problem and PCR primers in molecular dietary analysis: swamped or silenced; depth or breadth? Authorea.
Cuff et al. (2022). Evidence for nutrient-specific foraging of invertebrate predators under field conditions. Authorea.
Drake et al. (2021). Post-bioinformatic methods to identify and reduce the prevalence of artefacts in metabarcoding data. Authorea.
Badell-Grau and Cuff et al. (2020). Viral media: the prevalence of reactive online searching in the COVID-19 pandemic. Journal of Medical Internet Research Preprints.
Lafage et al. (2019). A new primer for metabarcoding of spider gut contents. PeerJ Preprints.
Protocols
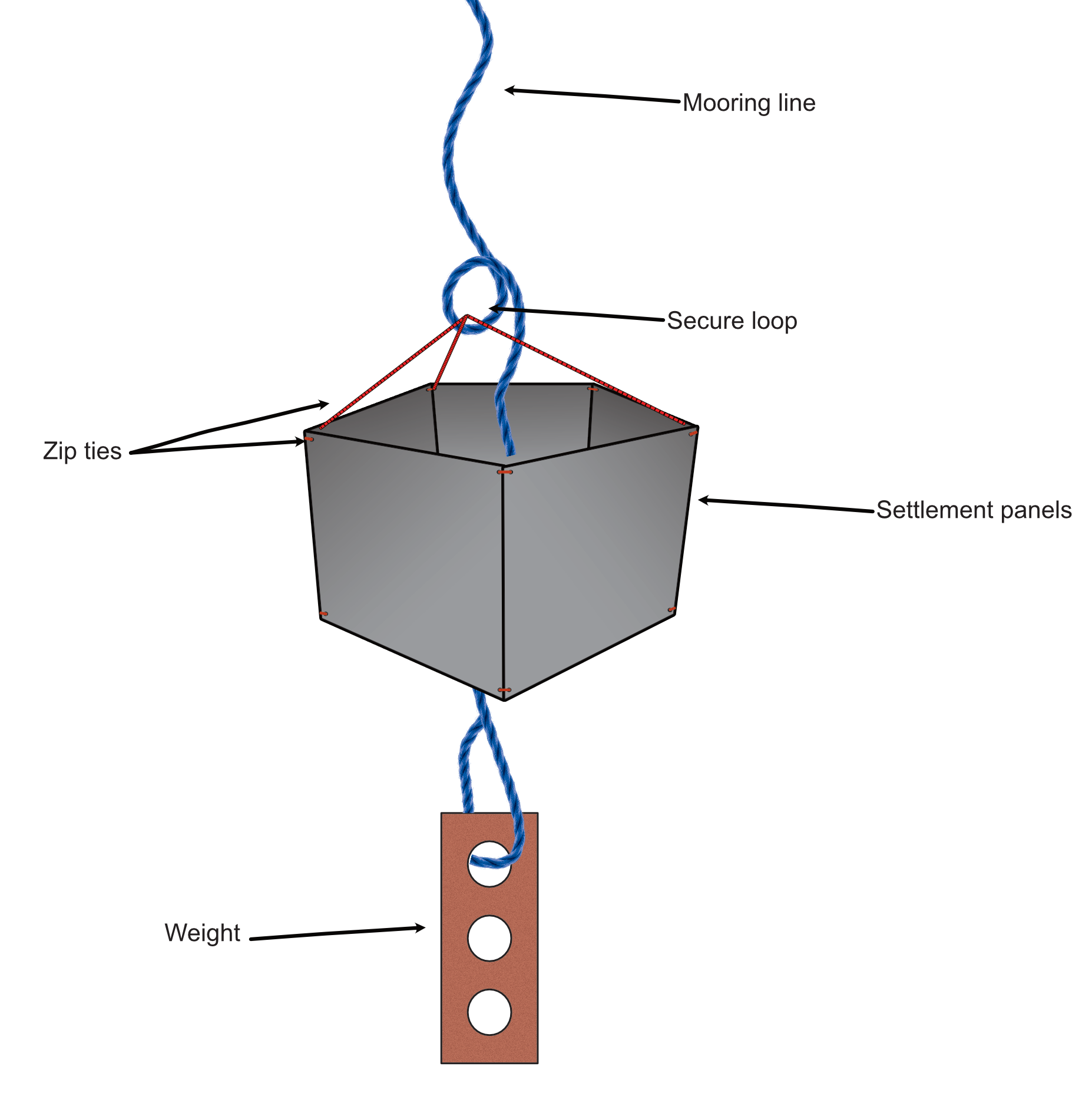
Longsden, Cuff & Sugden (2026). Molecular monitoring of biofouling communities using settlement panels and diagnostic assays. Protocols.io.
Cuff, Hawthorne, Christopher, Tercel & Evans (2025). Molecular analysis of trophic interactions for ecological network construction: EcoNet 2025 workshop. Protocols.io.
Cuff, Hawthorne, Maddock & Jensen (2025). Field-based invertebrate molecular diagnostics using a BentoLab. Protocols.io.
Tcheutchoua, Cuff & Sumner (2025). High-throughput wasp larvae gut content metabarcoding. Protocols.io.
Cuff, Kitson & Windsor (2025). Detecting arthropod leaf and flower visitation using DNA metabarcoding. Protocols.io.
Cuff, Howells, Kitson, Hawthorne & Evans (2025). High-throughput individual insect metabarcoding for identification and interaction data. Protocols.io.
Zaltz, Propistsova, Gavish-Regev & Cuff (2023). Metabarcoding based gut content analysis of arachnids. Protocols.io.

Cuff, Kitson & Evans (2023). Quantifying ecosystem service provider interactions via bulk sample DNA metabarcoding. Protocols.io.

Whittingham, Kitson & Cuff (2022). High-throughput papain-based DNA extraction from whole invertebrates. Protocols.io.

Cuff & Kitson (2022). High-throughput and cost effective pan trap DNA extraction. Protocols.io.

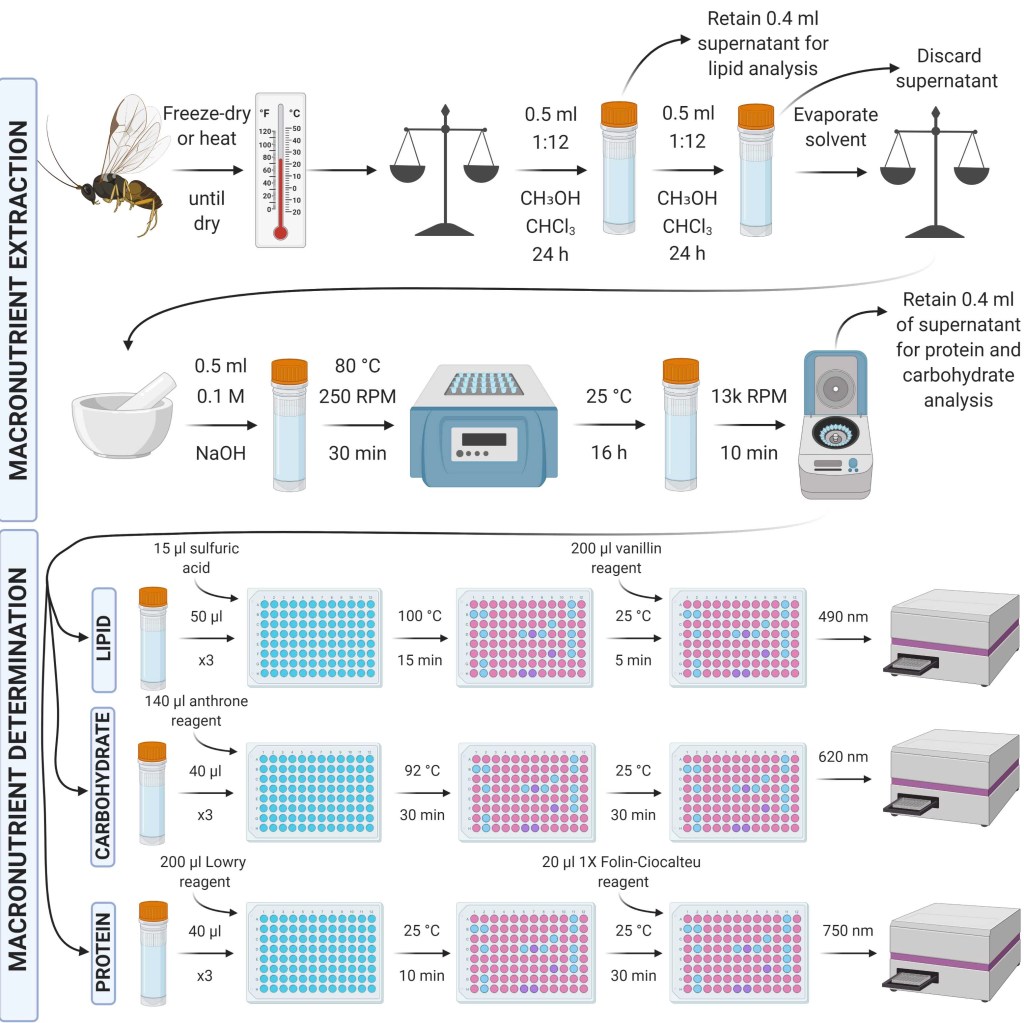
Cuff (2021). Further micro-scaled MEDI (Macronutrient Extraction and Determination from Invertebrates). Protocols.io.

Cuff & Wilder (2021). MEDI: Macronutrient Extraction and Determination from Invertebrates. Protocols.io.
Policy

Bruford et al. (2020). Cardiff University Ecosystem Resilience and Biodiversity Action Plan (ERBAP) 2021-2023. Cardiff University.

Bruford et al. (2019). Environment (Wales) Act 2016 Part 1 – Section 6: The Biodiversity and Resilience of Ecosystems Duty – Report 2019. Cardiff University.
Informal articles
Cuff & Tercel (2023). Obituary: Professor Bill Symondson. Antenna.
Cuff (2023). Rain or shine, spin and dine: unravelling how weather shapes spider food webs (and silk webs). Ecography Blog.
Cuff et al. (2022). Merging dietary metabarcoding into networks: turning “don’ts” into “dos”. Methods.blog.
Cuff & Drake (2022). Cover Stories: Cleaning up false positives with minimum sequence copy thresholds. Methods.blog.
Cuff and Tercel (2021). MEDI: Macronutrient Extraction and Determination from Invertebrates. Methods.blog.
Cuff (2020). British Arachnological Society Lockdown Spider Surveys. British Arachnological Society Newsletter.
Young and Cuff (2020). Four ways people stuck at home became armchair naturalists during lockdown. The Conversation.
Cuff and Rowson (2019). A Quick introduction to a prominent malacologist of the 20th Century. Mollusc World.

Cuff (2016). Home is where the heart-rot is. SEWBReC: Gwent-Glamorgan Recorders’ Newsletter.