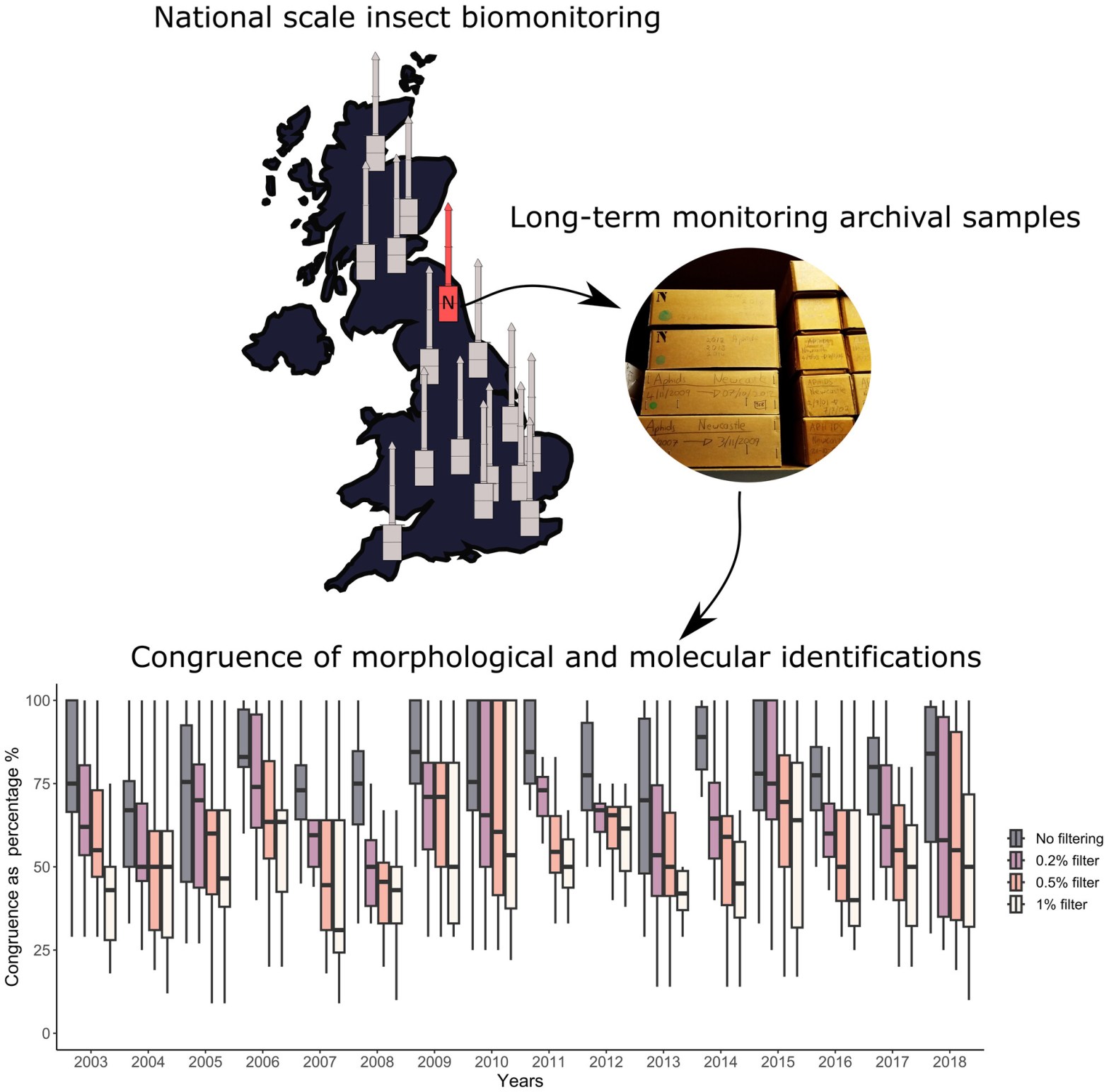
Check out this new paper published in Environmental DNA from Dimitrios Petsopoulos from the Network Ecology Group and co-authors: Identifying archived insect bulk samples using DNA metabarcoding: A case study using the long-term Rothamsted Insect Survey
Dimitrios led a study that demonstrates the compatibility of DNA metabarcoding with long-term insect monitoring schemes, using the Rothamsted Insect Survey as an example. Congruence between morphological and molecular datasets was variable, but didn’t relate to the time since collection, highlighting that long-term samples should provide a treasure trove of data for molecular ecological research. False positives and negatives are, however, a problem, and minimum sequence copy thresholds are not a fool-proof solution. Jordan really enjoyed helping out with the data interpretation for this, especially since it ties in nicely with a lot of other ongoing research adjacent to FERG in collaboration with the Network Ecology Group on biomonitoring.